
Our Work
The Furnari Lab studies malignant brain tumors across three core research areas: tumor heterogeneity, tumor dependencies and vulnerabilities, and translational brain tumor models. Our work examines how genetic and cellular diversity, including ecDNA organization, RNA modifications, and EGFR signaling, shapes tumor behavior and therapy resistance. In parallel, we uncover key molecular dependencies such as BRD2 signaling, PTEN-linked DNA repair weaknesses, and TERT-associated regulatory pathways, using clinically relevant patient-derived models to translate these discoveries toward improved brain cancer treatment.



Brett Taylor
Defining the Multi-Scale Organization and Regulation of Extrachromosomal DNA in Glioblastoma
My thesis work uses novel sequencing- and imaging-based genomic technologies to study the multi-scale regulation and organization of extrachromosomal DNA (ecDNA) in glioblastoma. Specifically, my project aims to answer two fundamental questions in the field: (1) What are the important biological features that mediate transcriptional regulation of ecDNA, and (2) what are the relationships between ecDNA, GBM cell identity, and tissue organization? By simultaneously measuring genomic alterations and transcription of single cells within their multi-scale spatial context, I hope to disentangle and clarify different facets of intratumoral heterogeneity, a fundamental obstacle in effective cancer therapy.
Dorothy Tsai
Identifying RNA Modifications and Their Effector Pathways in Glioblastoma Subtype Initiation, Progression, and Therapeutic Resistance.
There is emerging evidence that RNA modifications are linked to multiple cancers, but the specific modification types and proteins involved in GBM remains largely unknown. This project aims to map RNA modifications across GBM subtypes at single-cell resolution to discover subtype-specific signatures. By screening RNA modification writers, readers, and erasers associated with these signatures, we hope to identify pathways that regulate protein translation and GBM progression. This work will ultimately clarify the role of RNA modification programs in GBM to reveal subtype-specific epitranscriptomic therapeutic targets.

Clark Wang

Investigating ATRT cell of Origin and Tumor Heterogeneity via hiPSC-Derived Models
Atypical teratoid rhabdoid tumors (ATRTs) are rare, highly malignant pediatric brain cancers that almost always result from biallelic inactivation of SMARCB1. Despite presenting a remarkably simple genome defined by SMARCB1 loss, ATRTs are molecularly diverse, consisting of three subgroups with distinct DNA methylation profiles, transcriptomes, and clinical outcomes, suggesting differences in cells of origin and unique mechanisms of oncogenesis. Neural progenitor cells (NPCs) and neural crest cells (NCCs) have been proposed as potential cells of origin, with NPCs aligning more with the ATRT-SHH subgroup.
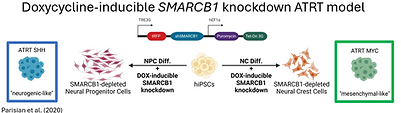
On the other hand, NCCs may account for the molecularly identical extracranial malignant rhabdoid tumors as well as the remaining intracranial subgroups, ATRT-TYR and ATRT-MYC, which have suspected extra-CNS origins. To study SMARCB1 loss in a genetically defined neural progenitor cellular context, our lab previously engineered human induced pluripotent stem cells (hiPSCs) with a doxycycline (DOX)-inducible SMARCB1 knockdown construct. NPCs derived from these hiPSCs, that were differentiated without SMARCB1 expression, exhibited an ATRT-SHH subgroup transcriptome and formed orthotopic tumors. Building upon these findings, these engineered hiPSCs were differentiated into neural crest cells to interrogate whether different cells of origin may contribute to the heterogeneity of ATRTs in patients.
Raghavendra Vadla, PhD
Epigenetic Plasticity and Therapeutic Resistance in Glioblastoma
My research focuses on defining how chromatin-modifying enzymes regulate mesenchymal state transitions, radiation resistance, and the emergence of drug-tolerant persister cells. I use integrated genetic, pharmacological, and multi-omics approaches, combining in vitro systems, CRISPR-based perturbations, and patient-derived xenograft models to uncover context-specific therapeutic vulnerabilities. My long-term goal is to translate these mechanistic insights into rational combination therapies that enhance treatment response and delay recurrence in aggressive brain tumors.
_edited.jpg)
Brandon Jones, PhD


Characterizing PTEN:Ki67 Interactions and Complex Contribution to Radioresistance in Glioblastoma
I study the tumor suppressor PTEN, an emerging player in GBM radioresistance .Advances in our understanding of GBM and decades of clinical trials have only had marginal improvement in survival outcomes. One hurdle to overcoming GBM recurrence and improving current standard of care therapy is the development of radioresistant tumors. More specifically, I investigate how PTEN interacts with Ki67 to upregulate DNA damage repair, preventing radiation induced DNA damage from killing tumor cells. Utilizing CRISPR/Cas9 in glioma stem-like cells, I characterize the changes in response to DNA damage in cells lacking Ki67 through various methodologies spanning cell proliferation assays, DNA damage repair foci quantification, and transcriptomic analysis. I also examine the physical binding of these 2 proteins using synthetic peptides and other biochemical techniques, with the goal of identifying unique binding properties which enable targeted disruption of this complex, serving as a potential launching point for radiosensitizing therapeutics in GBM.
Abhinaba Banerjee
Defining Biophysical Interactions Between EGFR Subclone Programs That Enhance Glioblastoma Invasion
My research investigates how intratumoral heterogeneity in receptor tyrosine kinase signaling drives glioblastoma (GBM) invasion. I focus on how interactions between genetically distinct tumor subclones with different EGFR states lead to bi-directional “education” that reprograms cell–matrix adhesion, mechanotransduction, and invasive behavior. Using integrated 2D, 3D, and in vivo platforms, including adhesion assays, spheroid invasion in brain-mimetic matrices, transcriptomic profiling, and whole-brain light-sheet imaging, I aim to define how signaling heterogeneity promotes cooperative tumor invasion. Ultimately, this work seeks to uncover biophysical and molecular vulnerabilities arising from tumor heterogeneity that can be leveraged to improve therapeutic strategies for GBM.




Christopher Chie
CRISPR screening in human iPSC-derived glioblastoma models reveals RNA-binding protein dependencies associated with TERT promoter mutation
Leveraging a stem cell–derived glioblastoma model and a high-throughput CRISPR screen, we aim to identify novel therapeutic vulnerabilities to target TERT promoter mutations, the most frequent genetic alteration in glioblastoma.
Takayuki Morimoto, MD
Harnessing Genome-edited Natural Killer cells for Immunotherapy of Glioblastoma Using Next generation models
My research focuses on understanding how tumor-intrinsic genetic alterations shape immune evasion and therapeutic resistance in glioblastoma (GBM). Using genetically engineered, murine NSC-derived GBM models, I investigate how defined driver muutations influence interactions between tumor cells and innate immune effectors, particularly natural killer (NK) cells. By integrating in vitro cytotoxicity assays, immune–tumor co-culture systems, and in vivo xenograft models, my work aims to identify tumor-specific mechanisms that regulate NK cell recognition, activation, and killing. Ultimately, this research seeks to uncover actionable immunotherapeutic vulnerabilities that can be exploited to enhance NK cell–based therapies and improve outcomes for patients with GBM.

Joseph Bendik
?????
??????
